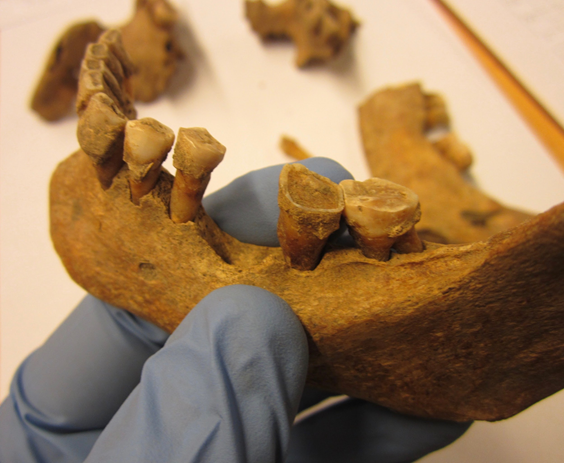
The communities of bacteria that live in our mouths have changed drastically since the Middle Ages, according to a new study of remains buried in a medieval Danish cemetery. And it turns out that some people may have been more predisposed to tooth and gum disease than others, thanks in part to the bacterial communities that lived in their mouths.
Ancient dental plaque
Biochemist Rosa Jersie-Christensen of the Novo Nordisk Foundation Center for Protein Research sampled hardened dental plaque, called calculus, from the skeletal remains of 21 Danish men who lived in the village of Tjærby between 1100 and 1450 CE. She and her colleagues chose men for the study because male immune systems tend to have stronger inflammatory responses, which would make it easier to find proteins associated with inflammation.
Overall, the men’s dental health wasn’t great—about what you might expect from a group of medieval villagers. All 21 showed some signs of gum disease, or periodontitis, along with at least minor cavities. Several had lost teeth sometime before their death.
The calculus had preserved thousands of proteins from the men’s bodies, their food, and the bacteria that lived in their mouths. Using mass spectrometry and a database of known proteins, Jersie-Christensen and her colleagues identified 3,761 different proteins in the samples. A few came from food, and others came from the host’s body. About 50 proteins came from blood plasma, and their presence suggests bleeding gums. But the majority of proteins—between 85 and 95 percent—had been produced by bacteria from the microbiome. There were proteins from about 220 different species in all.
Biochemist Enrico Cappellini of the Natural History Museum of Denmark, a coauthor on the study, says studying proteins can reveal which bacteria were active in the microscopic ecosystem of the mouth and how the host’s body responded to them. DNA can tell you which genes were present, but proteins tell you which genes were actually active.
“We were ultimately able to reconstruct which bacteria thrived in the mouth of medieval villagers and which physiological/pathological processes were going on there,” Cappellini told Ars Technica.
Not all microbiomes are created equal
All 21 men showed traces of bacteria known to cause gum disease and tooth decay. But the majority of proteins in samples from 16 of them came from several species of bacteria associated with a higher risk of tooth and gum disease. It’s probably no coincidence that nearly half of that group (seven men out of 16) showed signs of fairly severe tooth decay.
The second group had just one case of severe tooth decay. Cappellini says the proteins in those men’s dental plaque reflect more of a predisposition toward good health than those of their neighbors. Those proteins mostly came from relatively harmless bacteria, including Streptococcus sanguinis, a species that nearly always out-competes the main bacteria that causes dental cavities (its cousin Streptococcus mutans). That may help explain the relative lack of major cavities in the second group.
And the samples from that group also contained an abundance of human proteins associated with inflammation and other first-line responses to bacterial infection, as well as blood coagulation. Cappellini says the presence of so many of those proteins—which also turned up in large quantities in plaque from healthy modern volunteers—may mean that the second group had stronger immune responses, which could also have protected against more severe disease.
The two groups of men probably ate the same things, used the same (really bad) dental hygiene, and were exposed to the same pathogens. But the men in the first group had a microbiome that made them more vulnerable to infection, while the men in the second group may actually have gained some protection from their oral bacteria, Cappellini told Ars Technica.
We’ve come a long way
But despite their differences, all the medieval plaque samples had much more in common with each other than with the modern samples. While it’s possible that the medieval samples could be skewed in favor of the proteins most likely to survive a few hundred years of degradation, changes in lifestyle may account for most of the difference.
Today, people’s oral microbiomes vary much more from one person to the next than they did in the Middle Ages. The medieval men of Tjærby shared very similar lifestyles and monotonous diets; the traces of food proteins left in their dental plaque suggested dairy and goat milk, along with oats. In the 21st century, our diets include much more variety than the average medieval villager could have imagined, and one person’s diet may look much different from another’s. Different personal histories of antibiotic use, along with environmental factors and genetics, also shape microbial ecosystems in drastically different ways from person to person.
Scientists aren’t yet sure exactly how all of those factors combine to influence the invisible ecosystems that call our bodies home. In part, that’s because we don’t even completely understand which organisms live inside us; RNA studies have only recently discovered many species of oral bacteria, and several of them are nearly impossible to grow in the lab. But the sheer individuality of our microbiomes also presents a challenge. Some studies have suggested that the makeup of a person's microbiome may be as unique as a fingerprint, and that makes it hard to do large-scale comparisons of mouth microbiomes between populations.
But Jersie-Christensen and her colleagues say their study shows that general differences in bacteria populations can suggest something about a person’s overall oral health. And by combining studies of the proteins present in a person’s mouth with metagenomic studies of the microbiome, researchers may eventually be able to trace more connections between lifestyle, microbes, and health.
Nature Communications, 2018. DOI: 10.1038/s41467-018-07148-3 (About DOIs).